7.1 Predicting currents in the Openmanipulator joints with a neural network#
In this part we will use a neural network to predict the currents in our Openmanipulator robot arm. We will use real measured data of the currents and the joint angles of the first joint of our robot to train a neural network. First we will import the data and plot it to get a better understanding of the data. Secondly we will preprocess the data and train a neural network. Finally we will evaluate the performance of our neural network.
import Pkg # Package manager
# # Pkg.generate(joinpath(@__DIR__, "NeuronaleNetze")) # activate package environment
Pkg.activate(joinpath(@__DIR__, "NeuronaleNetze")) # activate package environment
Pkg.add("CSV") # add CSV
Pkg.add("Plots") # add Plots
Pkg.add("Statistics") # add Statistics
Pkg.add("ReverseDiff") # add ReverseDiff
Pkg.add("Flux") # add Machine Learning Library Flux
Pkg.instantiate()
Activating project at `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze`
Updating registry at `~/.julia/registries/General.toml`
Resolving package versions...
No Changes to `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze/Project.toml`
No Changes to `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze/Manifest.toml`
Precompiling project...
✓ GPUArrays
✓ Zygote
✓ Zygote → ZygoteColorsExt
✓ Flux
✓ NeuronaleNetze
5 dependencies successfully precompiled in 40 seconds. 213 already precompiled.
Resolving package versions...
No Changes to `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze/Project.toml`
No Changes to `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze/Manifest.toml`
Precompiling project...
✓ ColorVectorSpace → SpecialFunctionsExt
✓ ReverseDiff
✓ NeuronaleNetze
3 dependencies successfully precompiled in 8 seconds. 215 already precompiled.
Resolving package versions...
No Changes to `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze/Project.toml`
No Changes to `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze/Manifest.toml`
Resolving package versions...
No Changes to `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze/Project.toml`
No Changes to `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze/Manifest.toml`
Resolving package versions...
No Changes to `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze/Project.toml`
No Changes to `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze/Manifest.toml`
import Pkg # Package manager
Pkg.activate(joinpath(@__DIR__, "NeuronaleNetze")) # activate package environment
using CSV # CSV Parser
using Plots # Graph Plotter
using Statistics # statisitical functions, like "mean"
using Flux # Machine Learning Library
using ReverseDiff # Reverse Mode Automatic Differentiation
Activating project at `~/Library/Mobile Documents/com~apple~CloudDocs/Projects/numerical_methods/07-NeuralNetworks/NeuronaleNetze`
[ Info: Precompiling Flux [587475ba-b771-5e3f-ad9e-33799f191a9c]
[ Info: Precompiling SpecialFunctionsExt [997ecda8-951a-5f50-90ea-61382e97704b]
[ Info: Precompiling ZygoteColorsExt [e68c091a-8ea5-5ca7-be4f-380657d4ad79]
[ Info: Precompiling ReverseDiff [37e2e3b7-166d-5795-8a7a-e32c996b4267]
The Data#
Using the Openmanipulator we collected data using the Dynamixel Wizard tool which enables us to control and measure each actuator of our Robot. An image of the measured test data is shown below.
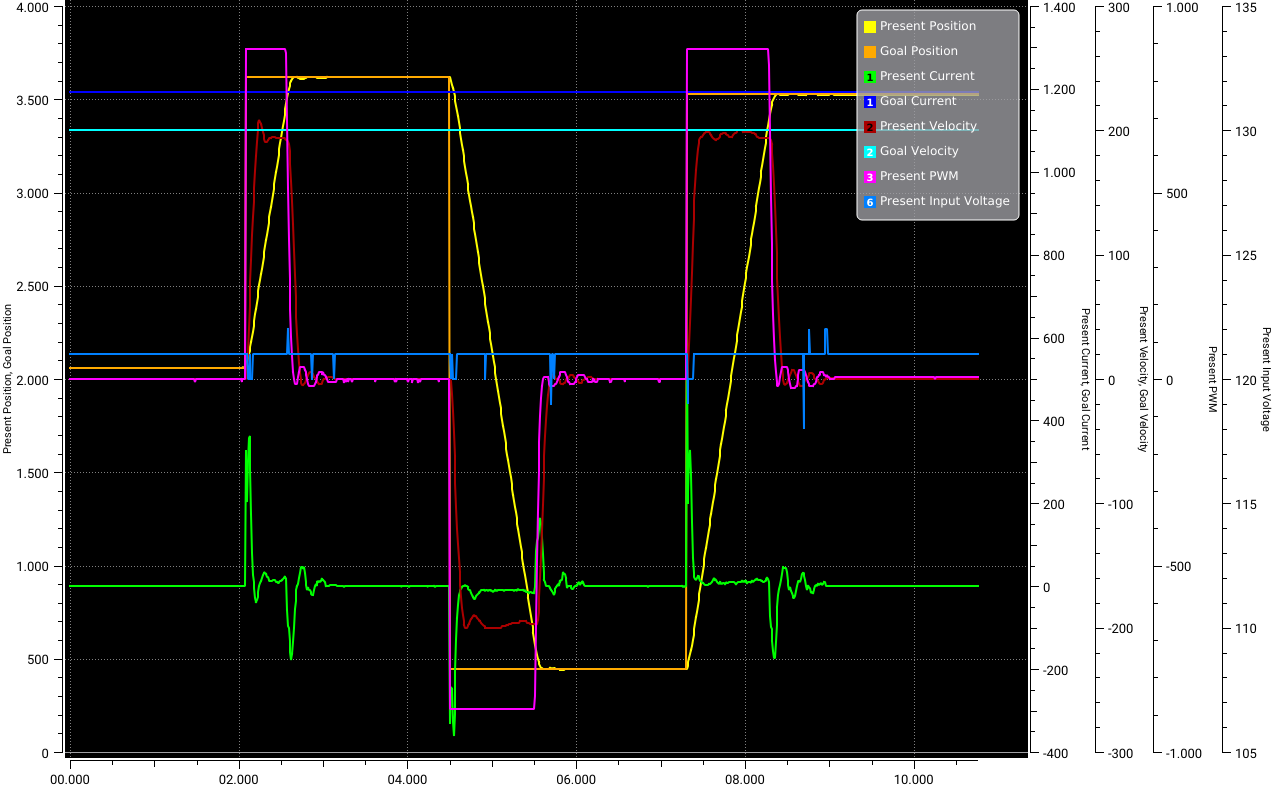
The data is stored in a CSV file. To load the CSV file, we can use the CSV module from Julia. For the input \(x\) into our model we will read the columns for:
Present Position
Goal Position
Present Velocity
We will make build the input matrix \(x\) of these values.
For the output \(y\) we will read the columns for:
Present Current
We will also make a matrix of these values. Since we are only predicting one value, we will only have one column. However, Flux expects a matrix, so we will make a matrix with one column.
function load_csv_data(filename)
csv = CSV.File(filename) # load and parse csv
x_1 = csv.columns[3].column # mask column 3 (Present Position) - input 1
x_2 = csv.columns[4].column # mask column 4 (Goal Position) - input 2
x_3 = csv.columns[7].column # mask column 7 (Present Velocity) - input 3
y = csv.columns[5].column # mask column 5 (Present Current) - this is what we want to predict
x_train = hcat(x_1, x_2, x_3)' # create input matrix
y_train = y # create output vector
y_train = reshape(y_train, 1, :) # reshape output vector to a matrix (1 x n) - this is what Flux expects
x_train, y_train
end
load_csv_data (generic function with 1 method)
# load training data
x_train, y_train = load_csv_data(joinpath(@__DIR__, "OMP_Daten/pc-train.csv"))
# load test data
x_test, y_test = load_csv_data(joinpath(@__DIR__, "OMP_Daten/pc-test.csv"))
([2061 2061 … 3525 3525; 2061 2061 … 3526 3526; 0 0 … 0 0], [0 0 … 0 0])
To normalize the data we estimate the mean and standard deviation of the data.
mean_x = mean(x_train)
mean_y = mean(y_train)
std_x = std(x_train)
std_y = std(y_train)
74.32507644633692
Task: Functions for preprocessing and postprocessing the data#
If we want the distribution of our data x to have a mean of \(\mu = 0\) and a standard deviation of \(\sigma = 1\) we can use the following:
Write a function to normalize the data using the mean and standard deviation.
Write a function to denormalize the data using the mean and standard deviation.
# data pre procseeing (make it machine frindly)
function dataPreProcess(x, y)
x = Float32.(x)
y = Float32.(y)
x = (x .- mean_x) ./ std_x
y = (y .- mean_y) ./ std_y
x, y
end
# data post procseeing (make it human frindly)
function dataPostProcess(x, y)
x = (x.* std_x) .+ mean_x
y = (y.* std_y) .+ mean_y
x, y
end
dataPostProcess (generic function with 1 method)
Preprocess the data using the function you wrote.
# Data Preprocessing
x_train, y_train = dataPreProcess(x_train, y_train)
([0.46310311404158183 0.46310311404158183 … 1.6074673134216337 1.6074673134216337; 0.46310311404158183 0.46310311404158183 … 1.6074673134216337 1.6074673134216337; -0.9706808124321389 -0.9706808124321389 … -0.9706808124321389 -0.9706808124321389], [-0.03509713558049676 -0.03509713558049676 … -0.04855154488368806 -0.03509713558049676])
Check the mean and std of the data to make sure we did it right.
@show mean(x_train)
@show std(x_train)
@show mean(y_train)
@show std(y_train);
mean(x_train) = 3.000783801098462e-17
std(x_train) = 1.0
mean(y_train) = 9.645376503530771e-18
std(y_train) = 1.0
We also need to normalize the test data. It is important to use the mean and std of the training data to normalize the test data since our neural network is trained on the normalized training data. Using a different mean and std for the test data would mean that we are using a different distribution for the test data than for the training data.
x_test, y_test = dataPreProcess(x_test, y_test)
([0.4736147264057587 0.4736147264057587 … 1.499548093149418 1.499548093149418; 0.4736147264057587 0.4736147264057587 … 1.50024886730703 1.50024886730703; -0.9706808124321389 -0.9706808124321389 … -0.9706808124321389 -0.9706808124321389], [-0.03509713558049676 -0.03509713558049676 … -0.03509713558049676 -0.03509713558049676])
Starting with a Linear Model#
In the lecture we showed that perceptrons can be build out of simple linear functions and adding a nonlinear activation layer. So before we will implement a neural network we take a step back and implement a linear function which estimates the currents.
The linear function is defined as:
where \(x\) is the input vector and \(w\) is the weight vector and \(b\) is the bias. We changed the notation to avoid adding the bias as a additional weight:
where \(w\) is now the extended weight vector and \(x\) is the input vector extended by a 1.
First lets look at an example of how to calculate the output of the linear function. We will use the first 3 rows of the training data as an example. The weight vector \(w\) is used to generate an output \(y\) for each row of the matrix \(X\):
x_1 = x_train[:, 1:3] # first 3 training examples each having 3 values
@show x_1 = vcat(ones(1, size(x_1, 2)), x_1)
@show w_1 = [1, 1, 1, 1]
@show x_1' * w_1;
x_1 = vcat(ones(1, size(x_1, 2)), x_1) = [1.0 1.0 1.0; 0.46310311404158183 0.46310311404158183 0.46310311404158183; 0.46310311404158183 0.46310311404158183 0.46310311404158183; -0.9706808124321389 -0.9706808124321389 -0.9706808124321389]
w_1 = [1, 1, 1, 1] = [1, 1, 1, 1]
x_1' * w_1 = [0.9555254156510247, 0.9555254156510247, 0.9555254156510247]
Task: Build a linear model#
Build a linear model using the definition we gave above.
# forward propagation
function linear_forward(x, w)
x = vcat(ones(1, size(x, 2)), x)
# Calculate the output of the neuron
y_hat = x' * w
end
# Define a function that takes in the weights and the data, and returns the loss
function loss(w, x, y)
y_hat = linear_forward(x, w)
mse = sum((y_hat .- y) .^ 2) / size(y, 1)
return mse
end
# Define a function that computes the gradients of the loss with respect to the weights
function grad(w, x, y)
return ReverseDiff.gradient(w -> loss(w, x, y), w)
end
# initialize model using random normal weights
function train_linear_model(x, y, learning_rate, num_epochs)
w = randn(4, 1)
mse = loss(w, x_t, y_t)
println("Start: MSE = $mse")
for i = 1:num_epochs
# Compute the gradients of the loss with respect to the weights
dw = grad(w, x, y)
# Update the weights using gradient descent
w -= learning_rate * dw
if i % 10 == 0
# Compute the loss and print it
mse = loss(w, x, y)
println("Iteration $i: MSE = $mse")
end
end
return w
end
train_linear_model (generic function with 1 method)
Before we start to train a linear model on the real dataset we will generate an artificial one where we know that we can actually learn it using a linear function. We will use the following to generate the data:
using Random
# Set the random seed for reproducibility
Random.seed!(1234)
# Define the true weights
w_true = randn(3, 1)
# Generate the dataset
n_samples = 1000
x_t = randn(3, n_samples)
y_t = x_t' * w_true
# Add some noise to the targets
y_t += 0.1 * randn(n_samples, 1);
w = train_linear_model(x_t, y_t, 0.01, 200)
@show w_true
@show w;
Start: MSE = 0.9464013338542072
Iteration 10: MSE = 0.6410760962735835
Iteration 20: MSE = 0.4353207509809646
Iteration 30: MSE = 0.2966379543575615
Iteration 40: MSE = 0.20314550700729128
Iteration 50: MSE = 0.14010606536329417
Iteration 60: MSE = 0.09759230842309466
Iteration 70: MSE = 0.06891573177013914
Iteration 80: MSE = 0.04956910738880395
Iteration 90: MSE = 0.03651453827477731
Iteration 100: MSE = 0.02770407606575343
Iteration 110: MSE = 0.021756869759651243
Iteration 120: MSE = 0.01774169161162987
Iteration 130: MSE = 0.015030417226521634
Iteration 140: MSE = 0.013199290987990745
Iteration 150: MSE = 0.011962379619805737
Iteration 160: MSE = 0.01112671182101594
Iteration 170: MSE = 0.010562031244568221
Iteration 180: MSE = 0.010180398681443182
Iteration 190: MSE = 0.00992243374207581
Iteration 200: MSE = 0.009748033165807008
w_true = [-0.3597289068234817; 1.0872084924285859; -0.4195896169388487;;]
w = [0.009873818517424525; -0.35469544923317287; 1.104683290673515; -0.4143009593658289;;]
Task: Train the linear model on the current dataset#
Now lots try to train the model on the real data. We already extracted the data from the CSV file and normalized it. So we can just start the training process by passing x_trainand y_train to the train_linear_model function.
train_linear_model(x_train, y_train, 0.01, 200)
Start: MSE = 7.291819186390243
Iteration 10: MSE = 7.76168422366377e128
Iteration 20: MSE = 4.730999090933705e249
Iteration 30: MSE = Inf
Iteration 40: MSE = Inf
Iteration 50: MSE = Inf
Iteration 60: MSE = NaN
Iteration 70: MSE = NaN
Iteration 80: MSE = NaN
Iteration 90: MSE = NaN
Iteration 100: MSE = NaN
Iteration 110: MSE = NaN
Iteration 120: MSE = NaN
Iteration 130: MSE = NaN
Iteration 140: MSE = NaN
Iteration 150: MSE = NaN
Iteration 160: MSE = NaN
Iteration 170: MSE = NaN
Iteration 180: MSE = NaN
Iteration 190: MSE = NaN
Iteration 200: MSE = NaN
4×1 Matrix{Float64}:
NaN
NaN
NaN
NaN
Ok, this does not seem to work. Can you think of a reason why?
Task: Build a linear model using Flux#
We can also build and train a linear model in a few lines of code using FLux. Build a linear model using Flux and train it on the data.
using Flux
# building a linear model using Flux
linear_model = Dense(3, 1)
# Define the loss function
loss(x, y) = sum((linear_model(x) .- y) .^ 2) / size(y, 1)
# Define the optimizer
# opt = Descent(0.01) # you can also try it with the ADAM optimizer
opt = ADAM(0.01)
# Train the model - check the train function and what we pass to it
# Note that the train function is called with train! which means that it will modify the model parameters passed to it
numIter = 200
for i in 1:numIter
Flux.train!(loss, Flux.params(linear_model), [(x_train, y_train)], opt, cb=() -> println(loss(x_train, y_train)))
end
┌ Warning: Layer with Float32 parameters got Float64 input.
│ The input will be converted, but any earlier layers may be very slow.
│ layer = Dense(3 => 1) # 4 parameters
│ summary(x) = "3×4420 Matrix{Float64}"
└ @ Flux ~/.julia/packages/Flux/pR3k3/src/layers/stateless.jl:60
14002.913710690085
13559.497538989885
13126.4067578622
12703.799507283413
12291.8187978055
11890.596126550869
11500.244824441706
11120.866801392363
10752.545886113761
10395.349424184522
10049.328495612432
9714.51726456128
9390.9310670419
9078.56687007451
8777.405330024443
8487.405959055548
8208.512430545221
7940.648237225319
7683.719897243034
7437.616823083517
7202.209822552375
6977.352913004886
6762.885434867702
6558.628476115785
6364.389894077638
6179.9623504808005
6005.125607611502
5839.647150626086
5683.282234250188
5535.776078369039
5396.864328227399
5266.273700755615
5143.724104691919
5028.929357604015
4921.597433216772
4821.432785217676
4728.137474312449
4641.411313353952
4560.953516408124
4486.4641765959805
4417.644560302683
4354.198683295788
4295.83418144344
4242.26321302579
4193.202832106652
4148.376722959518
4107.515206217871
4070.3561633768604
4036.645764166348
4006.1385843923167
3978.5988964407247
3953.800063580496
3931.5256082474048
3911.5691156623116
3893.7343508667127
3877.8356724930154
3863.6979469179605
3851.1562344233203
3840.0560566135628
3830.2531629647297
3821.613408140926
3814.0122373772338
3807.334775864467
3801.4752484493056
3796.3367689906063
3791.8309087321936
3787.877336716091
3784.403337995735
3781.3435133872954
3778.6392150330416
3776.2381916775266
3774.0941151314832
3772.1661613790843
3770.418583898764
3768.8203178369804
3767.34449815662
3765.968183209594
3764.671864988407
3763.439177557274
3762.256563875395
3761.112942860472
3759.999379749537
3758.9088798298712
3757.8360589904237
3756.7769391284983
3755.72872998986
3754.6896741928554
3753.6588304874585
3752.635880293135
3751.6210442861816
3750.6149750674695
3749.6185691829637
3748.6329839653035
3747.659465266551
3746.699304215482
3745.7538365727933
3744.8243042284203
3743.9119308398062
3743.017768830686
3742.142807744258
3741.2878713091422
3740.453635362819
3739.640667802013
3738.849293593003
3738.079828514175
3737.3323254960437
3736.606799359529
3735.9031016653717
3735.2209924493195
3734.5601322015914
3733.920102701145
3733.3004259190748
3732.700516827941
3732.1197637975165
3731.557574998967
3731.0132615510684
3730.4861104526635
3729.975465689017
3729.4806159205214
3729.0008602693033
3728.535536932689
3728.0839614309484
3727.6454608289077
3727.21944338715
3726.8052784507
3726.402424296002
3726.0102919868955
3725.6283593942862
3725.2561520123013
3724.8931704637525
3724.5390184217376
3724.1932255423185
3723.855477247499
3723.525351910371
3723.2025282993327
3722.8867099428567
3722.5775800193524
3722.2748624154074
3721.9782942529628
3721.6876450424834
3721.4026824567536
3721.123184602528
3720.8489890040855
3720.579837482975
3720.315563191356
3720.056076020969
3719.8011036689377
3719.5505444363234
3719.3042514203134
3719.0620574711693
3718.823858344658
3718.5894562994886
3718.358791119743
3718.131713739569
3717.9081093618333
3717.6878565021516
3717.4708564289485
3717.256973698428
3717.04614688369
3716.8382435215763
3716.633184473755
3716.430865423321
3716.231190703256
3716.0340904227037
3715.8394595583586
3715.647228007112
3715.4573369656027
3715.2696896816597
3715.0842207736932
3714.9008617556206
3714.7195574035054
3714.540225323823
3714.3628185073285
3714.187275168858
3714.013535777336
3713.841578046593
3713.671292226722
3713.50268391279
3713.335705164678
3713.170284571069
3713.0064070646467
3712.8440110589654
3712.6830757713697
3712.5235486923457
3712.365403496533
3712.2086419850475
3712.053185494183
3711.8990274371636
3711.74612560319
3711.5944686445896
3711.4440229583183
3711.29475215343
3711.1466610736225
3710.9997146124683
3710.853881714872
3710.709160005305
3710.565504535969
3710.42294156131
3710.2813945398366
3710.14087922401
Linear model with nonlinear activation function using Flux#
Now we extend the linear model by adding a nonlinear activation function. We will use the sigmoid function as activation function using Flux.sigmoid. Our function is then defined as:
where \(h\) is the sigmoid function.
# build linear model with nonlinear sigmoid activation function
non_linear_model = Dense(3, 1, Flux.tanh)
# Define the loss function
loss(x, y) = sum((non_linear_model(x) .- y) .^ 2) / size(y, 1)
# Define the optimizer
opt = Descent(0.01) # you can also try it with the ADAM optimizer
# Train the model
numIter = 200
for i in 1:numIter
Flux.train!(loss, Flux.params(non_linear_model), [(x_train, y_train)], opt, cb=() -> println(loss(x_train, y_train)))
end
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
8839.000000000004
Neural Network#
Now we have seen that we cannot learn the current of the actuators using a linear model. We also adapted the linear model with a nonlinear activation function. Now we will use several layers of these linear functions with nonlinear activation function to build a feed forward neural network. We will use a fully connected feed forward neural network with 1-2 hidden layers and a linear output layer. To build such a fully connected feed forward neural network we can use the chain function from Flux. The chain function takes a list of layers as input and builds a neural network with these layers. In this case we will use Dense layers which are fully connected layers. The Dense layer takes the number of input neurons, the number of output neurons and the activation function as input.
Task: Build the neural network#
Build a neural network with 1-2 hidden layer. The input layer should have 3 neurons and the output layer should have 1 neuron to fit our data. You can also test different activation functions and different numbers of neurons in the hidden layer.
You can check the different activation function in flux here: https://fluxml.ai/Flux.jl/stable/models/activation/
To optimize the neural network we will use the ADAM optimizer and set its learning rate to \(0.001\). You can check the different optimizers in flux here: https://fluxml.ai/Flux.jl/stable/training/optimisers/
# define our Neural Network model
function build_model()
opt = ADAM(0.001)
input_size = size(x_train)[1]
output_size = size(y_train)[1]
model = Chain(Dense(input_size, 32, relu),
Dense(32, 32, relu),
Dense(32, output_size, identity))
model, opt
end
build_model (generic function with 1 method)
Task: define the forward function#
We also define a forward function to calculate the output of the neural network.
function forward(x)
y_hat = model(x)
end
forward (generic function with 1 method)
Task: define the loss function#
We also need a loss function to optimize. Since we are doing regression, we will use the mean squared error loss function. You can check the different loss functions in flux here: https://fluxml.ai/Flux.jl/stable/models/losses/
# Mean square error as the loss function for optimization
function loss(y_hat, y)
sum((y_hat .- y) .^ 2)
end
loss (generic function with 2 methods)
Task: Train the neural network#
Now we will put everything together and train the neural network. We will define a function which takes the neural network, the optimizer, and the number of epochs as input. The function should return the loss based on the training and test data.
# train the model
function train_model(model, opt, numIter)
trainLoss = zeros(numIter)
testLoss = zeros(numIter)
opt_state = Flux.setup(opt, model)
for i in 1:numIter
Flux.train!(model, [(x_train, y_train)], opt_state) do m, x, y
loss(m(x), y)
end
trainLoss[i] = loss(forward(x_train), y_train) / length(y_train)
testLoss[i] = loss(forward(x_test), y_test) / length(y_test)
end
trainLoss, testLoss
end
train_model (generic function with 1 method)
Calling the build_model() and train_model() functions:
model, opt = build_model() # build model
trainLoss, testLoss = train_model(model, opt, 1000) # train model
([1.0404954178824837, 1.0285629720485543, 1.017815951617186, 1.0082485489397477, 0.9997252074294303, 0.9920813242593242, 0.9851829202102648, 0.9788624355497945, 0.972989111670157, 0.9674405189160014 … 0.2174361180080419, 0.217370944007769, 0.21728451948155164, 0.2171750755469239, 0.21704500367468335, 0.2169052891127391, 0.21676045550053588, 0.216631430784194, 0.2165241132571936, 0.21644143808898125], [0.48593526444818375, 0.4748848904272836, 0.4652117723717658, 0.457062131596801, 0.4502811758449016, 0.4445673953083006, 0.43978542397853015, 0.435733750980032, 0.43217385460098906, 0.4288505615250615 … 0.08996966574812512, 0.09186244920673922, 0.08975519515549303, 0.0916835265351677, 0.08970905910395766, 0.0912221726020237, 0.08991361642104072, 0.09048910576676693, 0.09035815498838777, 0.08987093449581995])
Plotting the train and test loss:
# plot the train loss
plot(trainLoss, label="Train Loss", xlabel="Iterations", ylabel="loss")
# plot the test loss
plot!(testLoss, label="Test Loss", xlabel="Iterations", ylabel="loss")
Let’s also plot the predicted currents and the actual currents based on the training data:
# plot the results for the training data
y_hat = forward(x_train)
plot(y_train[1, :], label="Ground Truth (train)", ylabel="Current")
plot!(y_hat[1, :], label="Prediction (train)")
Let’s do the same for the test data:
# test the final model on the test data
y_hat = forward(x_test)
error = sum((y_hat .- y_test) .^ 2) / length(y_test)
println("Test Error: ", error)
# post process the data (make it human readable)
x_test, y_test = dataPostProcess(x_test, y_test)
_, y_hat = dataPostProcess(x_test, y_hat)
# plot the results
plot(y_test[1, :], label="Ground Truth (test)", ylabel="Current")
plot!(y_hat[1, :], label="Prediction (test)")
# error in human readable form
error = y_hat .- y_test
# plot the error
plot!(error[1, :], label="Error", ylabel="Error")
Test Error: 0.08987093449581995
Could we replace a simulation with a neural network?#
In this part we will investigate how well a neural network can predict the change of the joint angle of the first joint of the Openmanipulator-X based on the joint angle, the goal joint angle and the current in the actuator.
function load_csv_data(filename)
csv = CSV.File(filename) # load and parse csv
x_1 = csv.columns[3].column[1:end-1] # mask column 3 (Present Position)
x_2 = csv.columns[4].column[1:end-1] # mask column 4 (Goal Position)
x_3 = csv.columns[5].column[1:end-1] # mask column 5 (Present Current)
# the prediction will be the delta position of the joint (Present Position at time t+1 - Present Position at time t)
y = csv.columns[3].column[2:end] - csv.columns[3].column[1:end-1] # mask column 3 (delta Position) - this is what we want to predict
x_train = hcat(x_1, x_2, x_3)' # create input matrix
y_train = y # create output vector
y_train = reshape(y_train, 1, :) # reshape output vector to a matrix (1 x n) - this is what Flux expects
x_train, y_train
end
load_csv_data (generic function with 1 method)
# load training data
x_train, y_train = load_csv_data(joinpath(@__DIR__, "OMP_Daten/pc-train.csv"))
# load test data
x_test, y_test = load_csv_data(joinpath(@__DIR__, "OMP_Daten/pc-test.csv"))
([2061 2061 … 3525 3525; 2061 2061 … 3526 3526; 0 0 … 0 0], [0 0 … 0 0])
Lets plot the training data to get a better understanding of the data:
# plot the data and set the heading
p1 = plot(x_train[1, :], label="P-Pos", ylabel="x_1")
p2 = plot(x_train[2, :], label="G-Pos", ylabel="x_2")
p3 = plot(x_train[3, :], label="P-I", ylabel="x_3")
p4 = plot(y_train[1, :], label="∂-Pos", ylabel="y")
plot(p1, p2, p3, p4, layout=(4, 1))
Normalize the data:
# calculate mean and standard deviation of the training data
mean_x = mean(x_train)
mean_y = mean(y_train)
std_x = std(x_train)
std_y = std(y_train)
# Data Preprocessing
x_train, y_train = dataPreProcess(x_train, y_train)
x_test, y_test = dataPreProcess(x_test, y_test)
([0.4741199295295133 0.4741199295295133 … 1.5008998289755333 1.5008998289755333; 0.4741199295295133 0.4741199295295133 … 1.5016011813658654 1.5016011813658654; -0.9713673469446992 -0.9713673469446992 … -0.9713673469446992 -0.9713673469446992], [-0.02003504923446377 -0.02003504923446377 … -0.02003504923446377 -0.02003504923446377])
Now we build the model and train it. This time we will train it for \(20000\) epochs. The rest of the parameters stay the same.
model, opt = build_model() # build model
trainLoss, testLoss = train_model(model, opt, 20000) # train model
([1.0228227214352001, 0.9965736005250969, 0.9713614819069315, 0.9472253978897653, 0.9241210379544437, 0.9019925339522422, 0.8807524413520813, 0.8603005902150264, 0.840594735329256, 0.8215905536316962 … 0.004471555799321393, 0.004474152789156546, 0.004476836473598991, 0.004480359682074878, 0.0044837654904354715, 0.004488161261447957, 0.00449241915864023, 0.00449713085261265, 0.004501766470505711, 0.004508047973317812], [0.6864295240501983, 0.6645025907139858, 0.6438979845255944, 0.6246338660548105, 0.6066765100605235, 0.5899084748280882, 0.5742130991294334, 0.5594753129784247, 0.545688795919504, 0.5327951963422685 … 0.002043555236727043, 0.0020900571115619925, 0.002050709934658085, 0.002102025327968878, 0.0020542891208735403, 0.0021171765520147665, 0.0020624494016207727, 0.00213532530581838, 0.002070472361886978, 0.002153709882977916])
Let’s plot the train and test loss:
# plot the train loss
fig = plot(1:numIter, trainLoss, label="Train Loss", xlabel="Iterations", ylabel="loss")
# plot the test loss
plot!(1:numIter, testLoss, label="Test Loss", xlabel="Iterations", ylabel="loss")
Check how well the model predicts the joint angle change based on the training data:
# plot the results for the training data
y_hat = forward(x_train)
plot(y_train[1, :], label="Ground Truth (train)", ylabel="Position")
plot!(y_hat[1, :], label="Prediction (train)")
Let’s do the same for the test data:
# test the final model on the test data
y_hat = forward(x_test)
error = sum((y_hat .- y_test) .^ 2) / length(y_test)
println("Test Error: ", error)
# post process the data (make it human readable)
x_test, y_test = dataPostProcess(x_test, y_test)
_, y_hat = dataPostProcess(x_test, y_hat)
# plot the results
plot(y_test[1, :], label="Ground Truth (test)", ylabel="Position")
plot!(y_hat[1, :], label="Prediction (test)")
# error in human readable form
error = y_hat .- y_test
# plot the error
plot!(error[1, :], label="Error")
Test Error: 0.002153709882977916
